Tutorial
Welcome to the DeepRetro tutorial! This section focuses on using the DeepRetro GUI for Retrosynthesis Analysis and Pathway Visualization
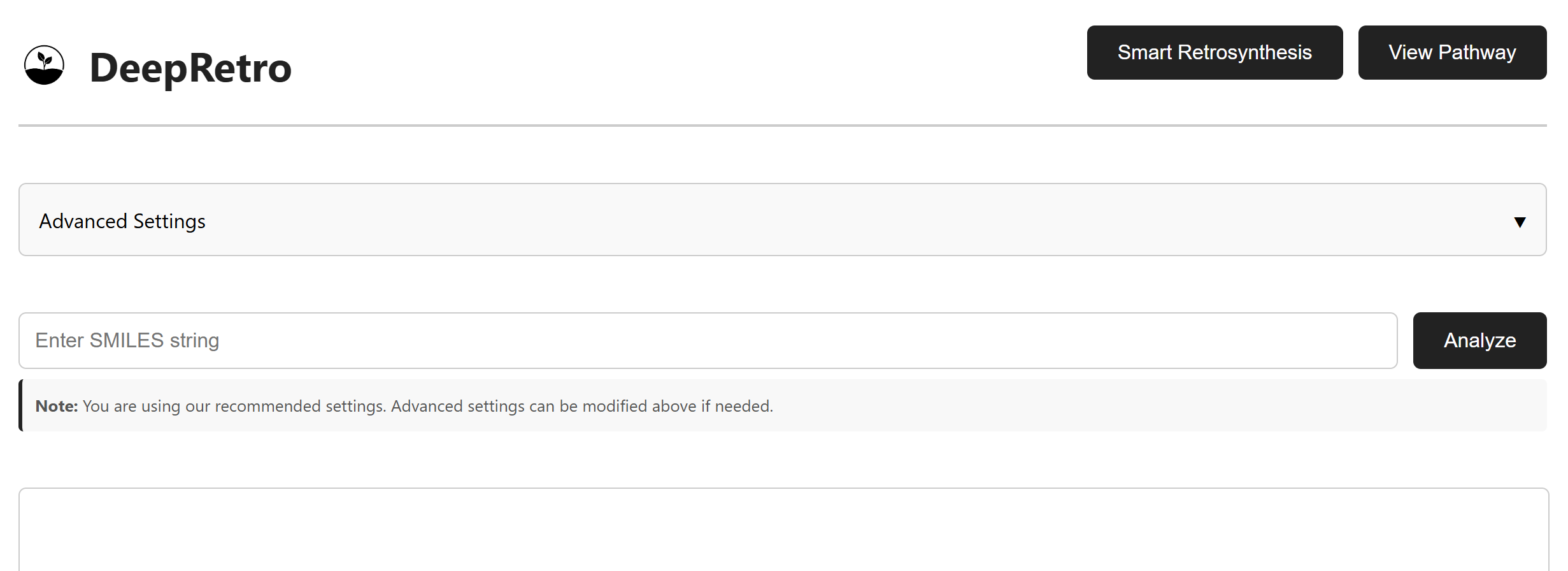
Getting Started
Prerequisites
You should have the DeepRetro backend server running
You should have an API key for backend access
Try using a modern web browser (Chrome/Firefox/Safari)
Setup
Navigate to viewer/index.html
Enter your frontend API key when prompted
Choose your analysis mode (Smart Retrosynthesis or View Pathway)
Advanced Settings Configuration
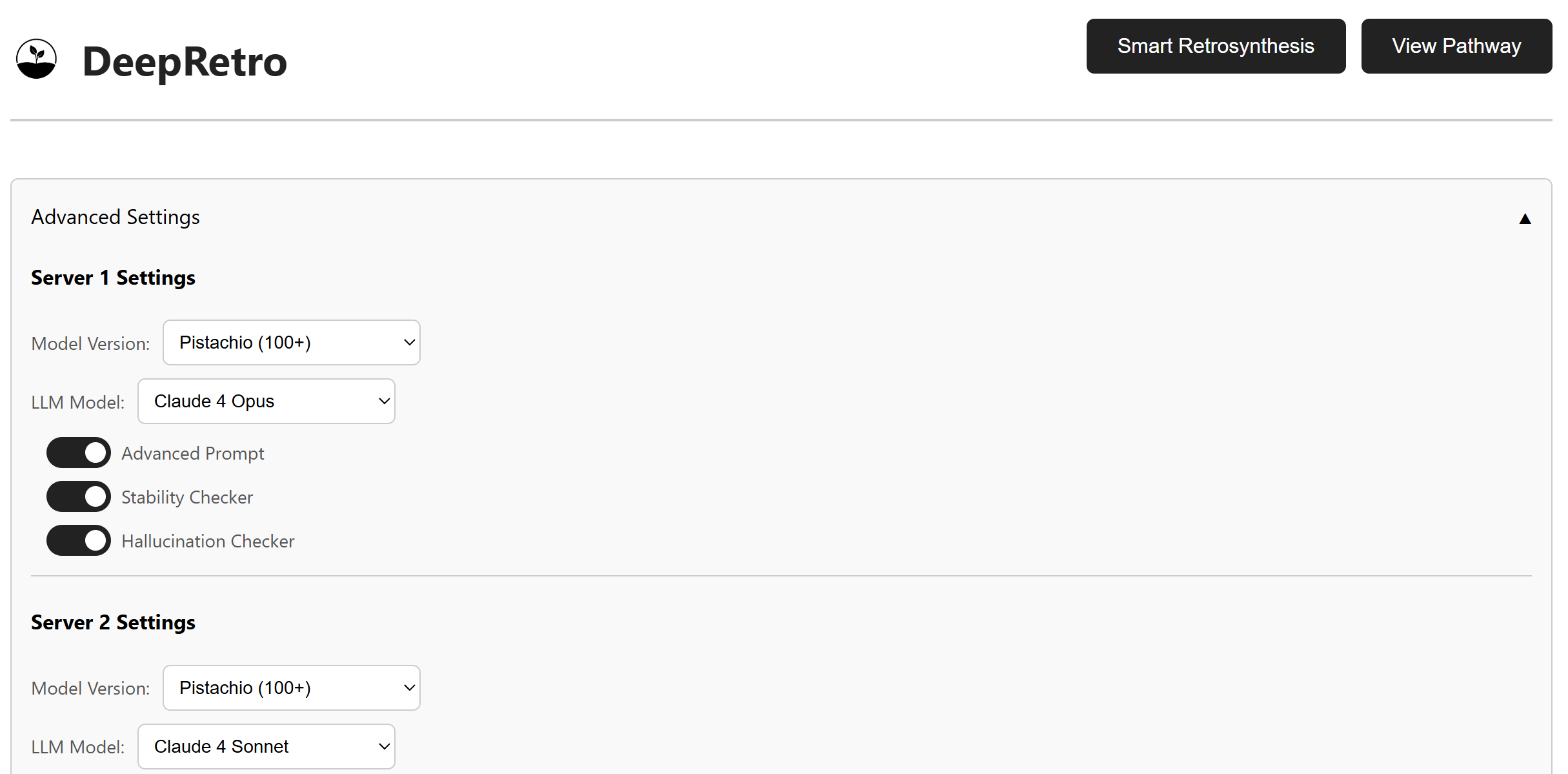
The advanced settings panel consists of multiple options that can be configured to optimize the retrosynthesis process. While the default settings are optimum, the LLM model, backend model, and validation checks can be adjusted to suit your needs.
LLM Selection:
Claude 3 Opus, Claude 3.7 Sonnet, DeepSeek, Claude 4 Opus, Claude 4 Sonnet
Model Backend Selection:
USPTO, Pistachio_25, Pistachio_50, Pistachio_100, Pistachio_100+
Validation Checks:
Stability validation enable/disable, Hallucination detection settings, Chemical feasibility assessment
SMILES Input and Configuration
Target Molecule Entry
The SMILES input interface allows you to enter the target molecule for retrosynthesis analysis. Here we are using Cyanostilbene as an example:
"COc1ccc(-c2ccc(/C=C(\\C#N)c3ccc(-c4ccncc4)cc3)cc2)cc1"
We then click “Analyze” to start the retrosynthesis process and the system will validate the SMILES and begin pathway generation.
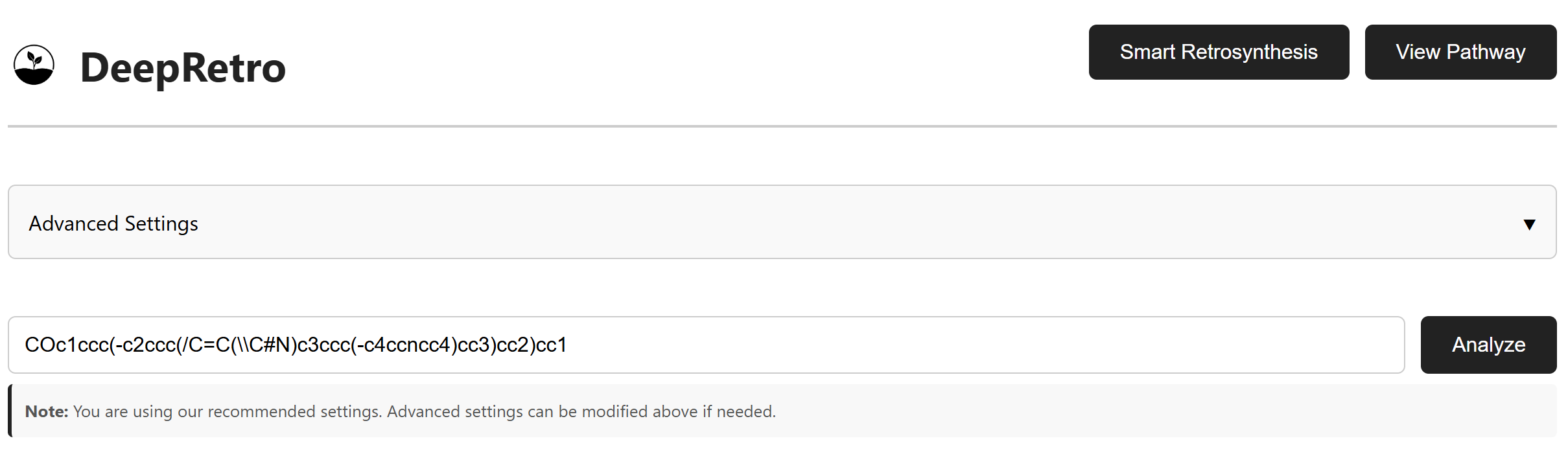
Input Requirements
Please make sure you are using a valid SMILES notation (Simplified Molecular Input Line Entry System) and have a chemically feasible target molecule.
Visualization
Pathway Visualization
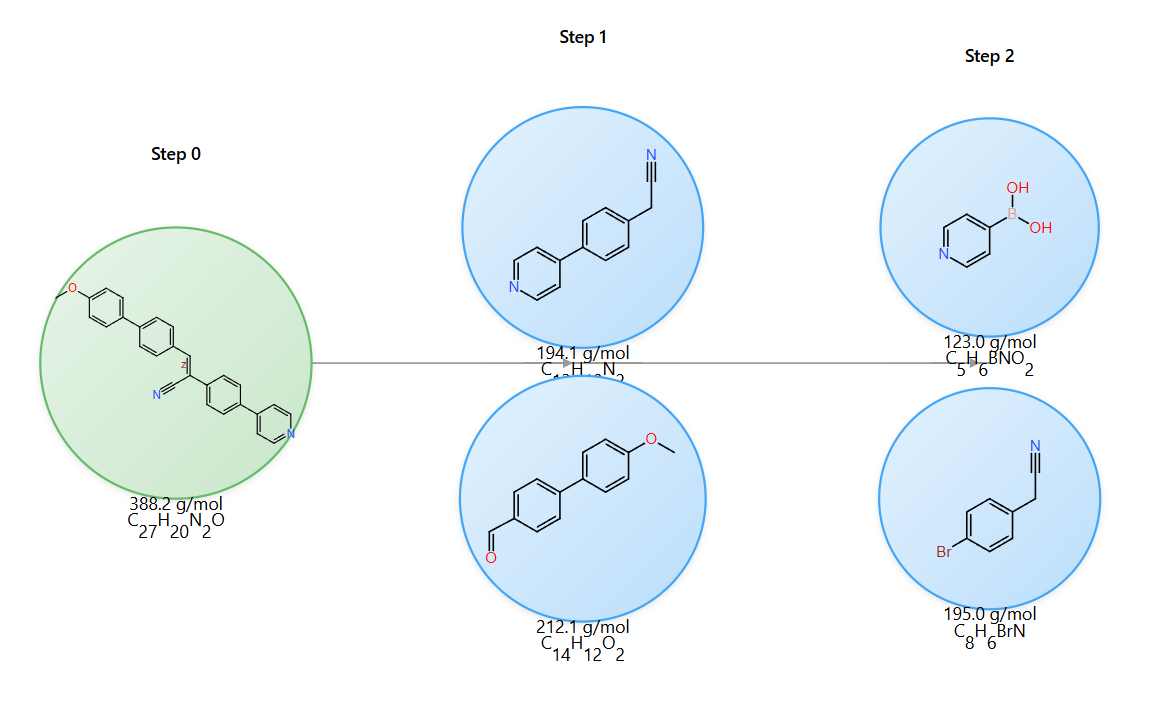
Chemical Context: Cyanostilbene Synthesis
The example demonstrates synthesis of a cyanostilbene derivative - a valuable chromophore with aggregation-induced emission (AIE) and strong donor–acceptor charge transfer properties.
- Two-Step Synthesis Approach
Step 1 (Knoevenagel Condensation): Introduces the cyano group, forming the final conjugated system
Step 2 (Suzuki Coupling): Connects the donor and acceptor aromatic units
Molecular Information
The visualization provides several interactive elements to help you explore and analyze retrosynthesis pathways. When examining molecule nodes, you can hover on any node to view detailed structural information, including molecular weight, chemical formula, and access the SMILES notation along with the step metrics.
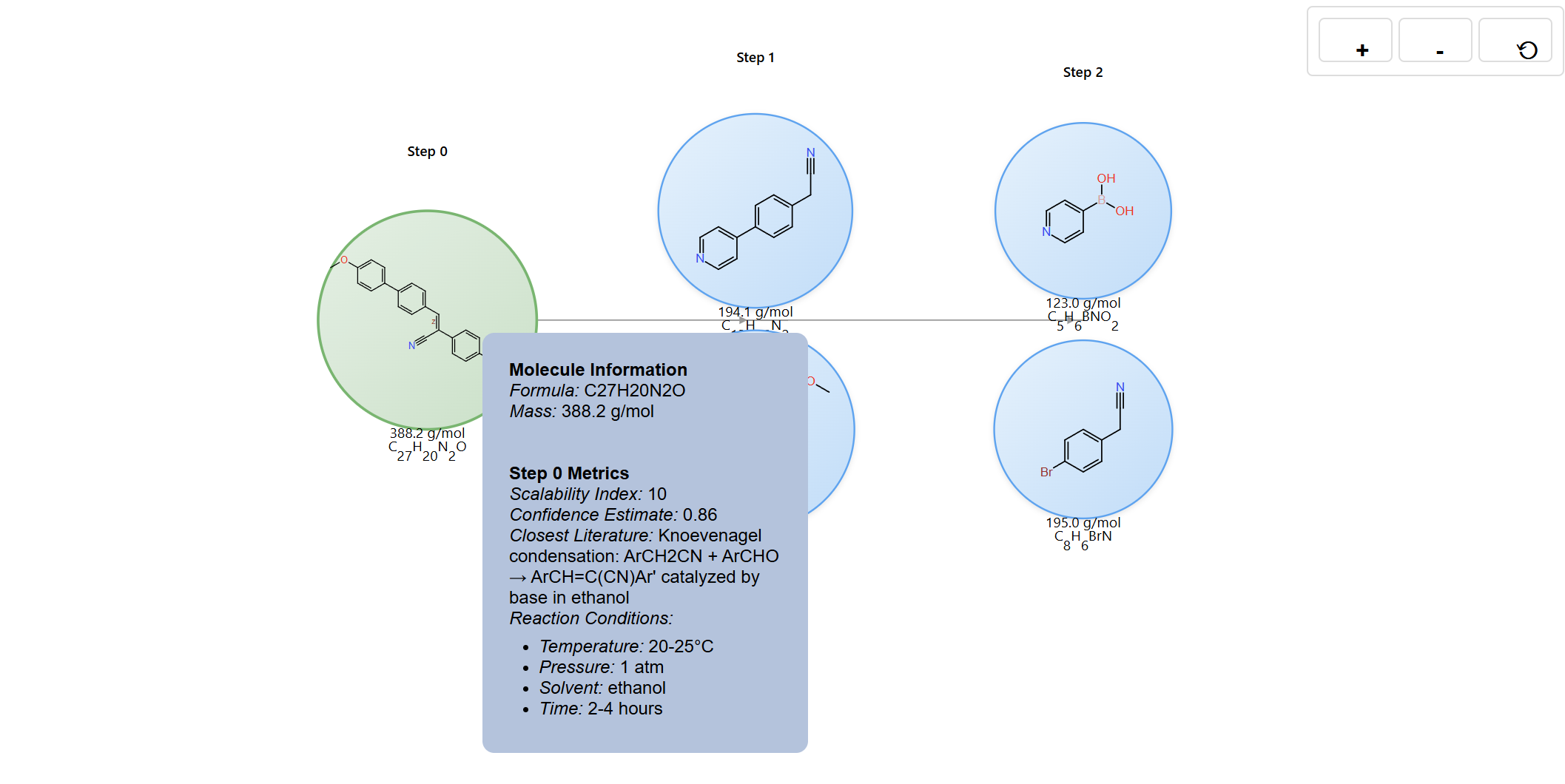
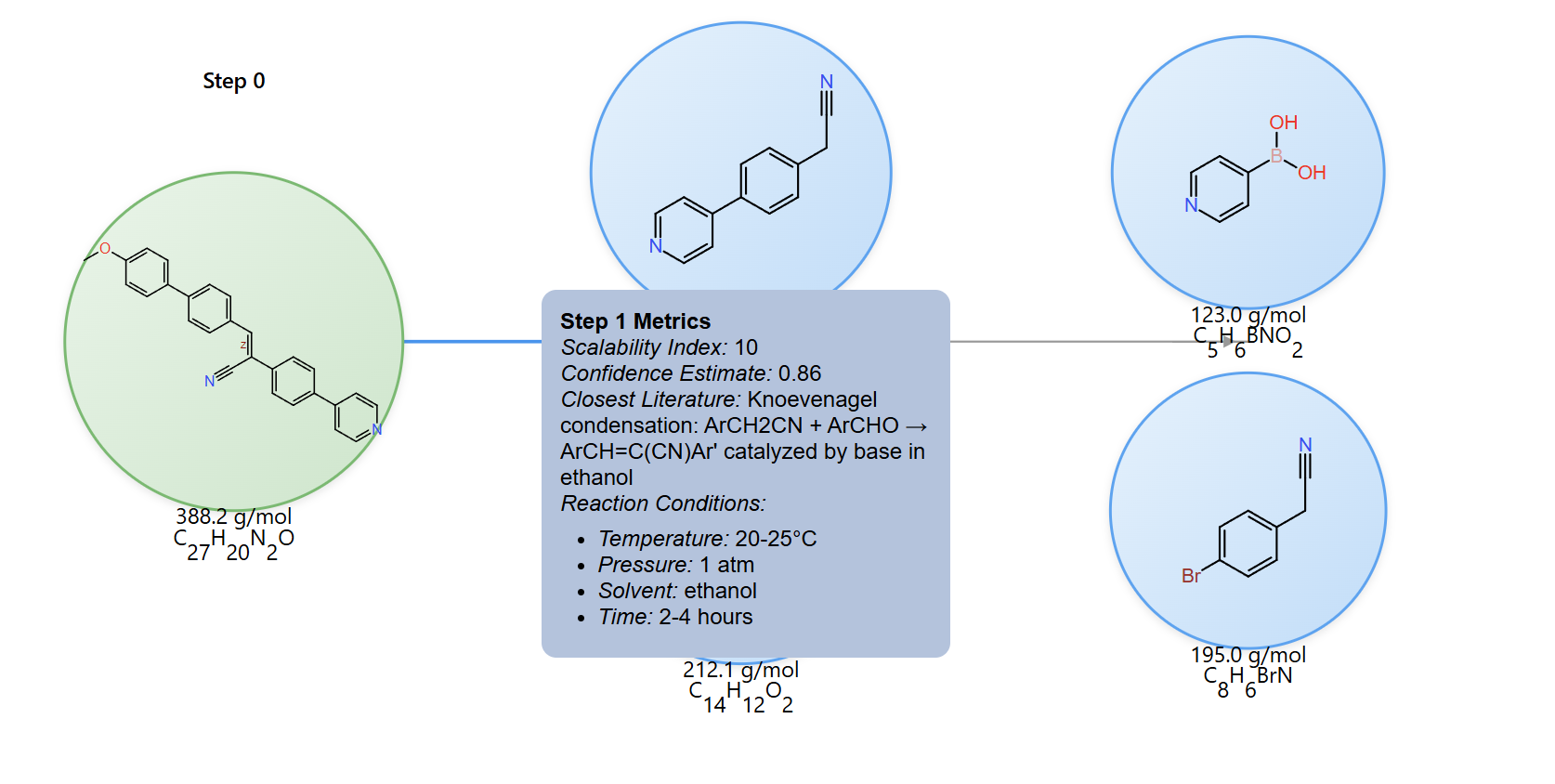
For reaction edges connecting the molecules, hover your mouse over them to reveal important reaction conditions and metrics. This includes the scalability index, confidence estimate and closest literature metadata along with reaction conditions such as temperature and solvent information.
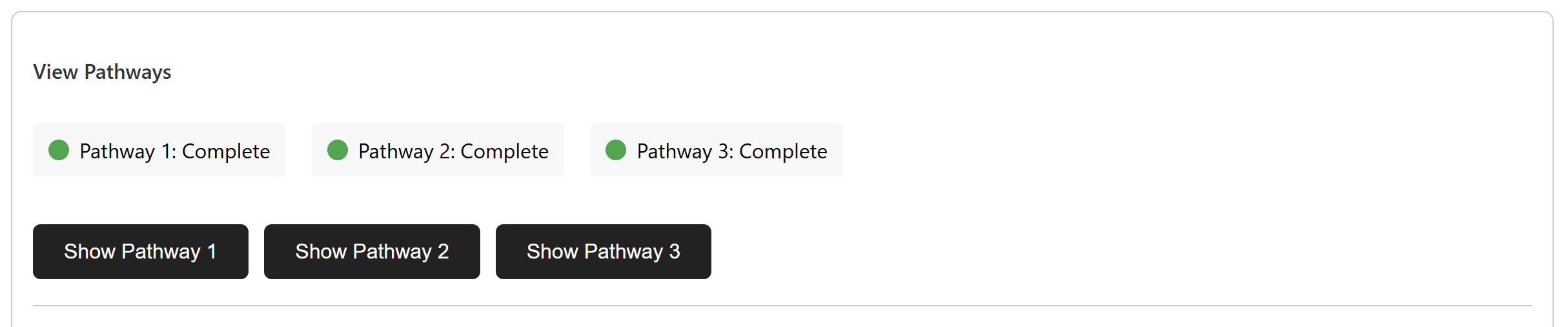
To effectively navigate complex reaction networks, the interface offers several navigation tools. You can easily switch between different proposed pathways, zoom in and out to examine specific details, and pan across larger reaction trees. The step-by-step progression view allows you to follow the synthesis route sequentially, while the comparison feature lets you evaluate multiple pathways side by side to determine the optimal route.
Interactive Editing
Partial Re-run Analysis
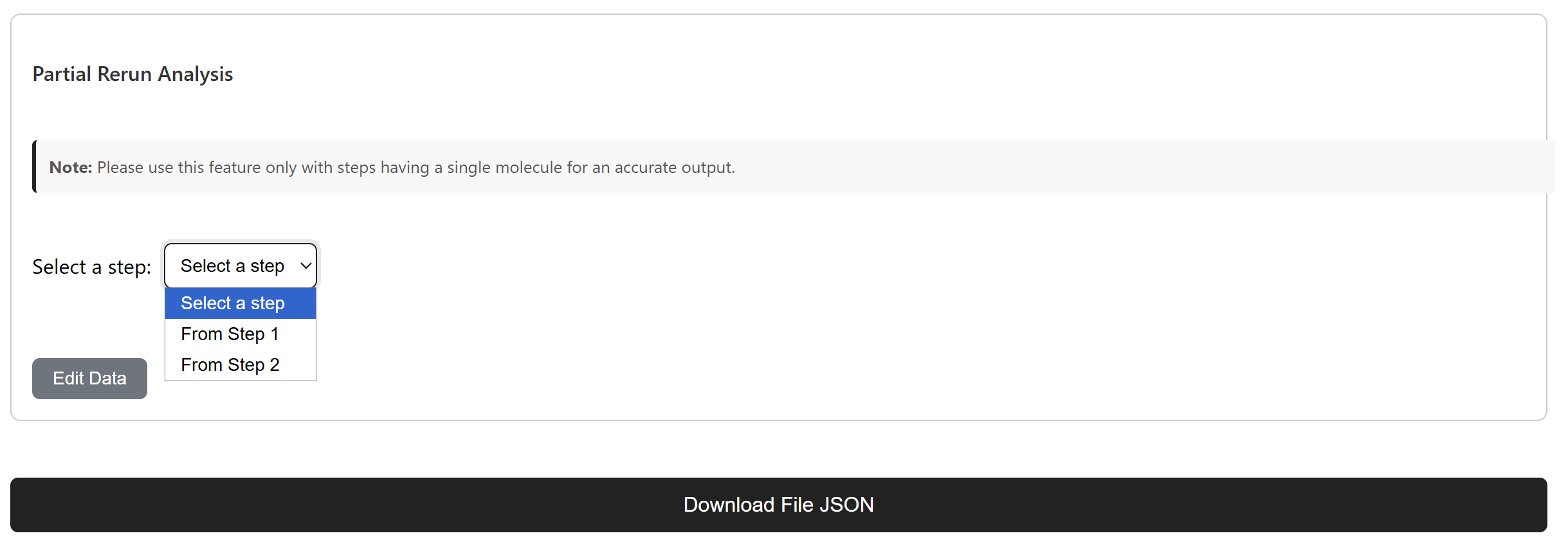
- Expert chemist intervention workflow:
The expert chemist can select a specific reaction step that needs modification and generate new pathway branches starting from that modified step. This allows for targeted refinement of problematic reactions while preserving successful parts of the route.
Manual Pathway Editing
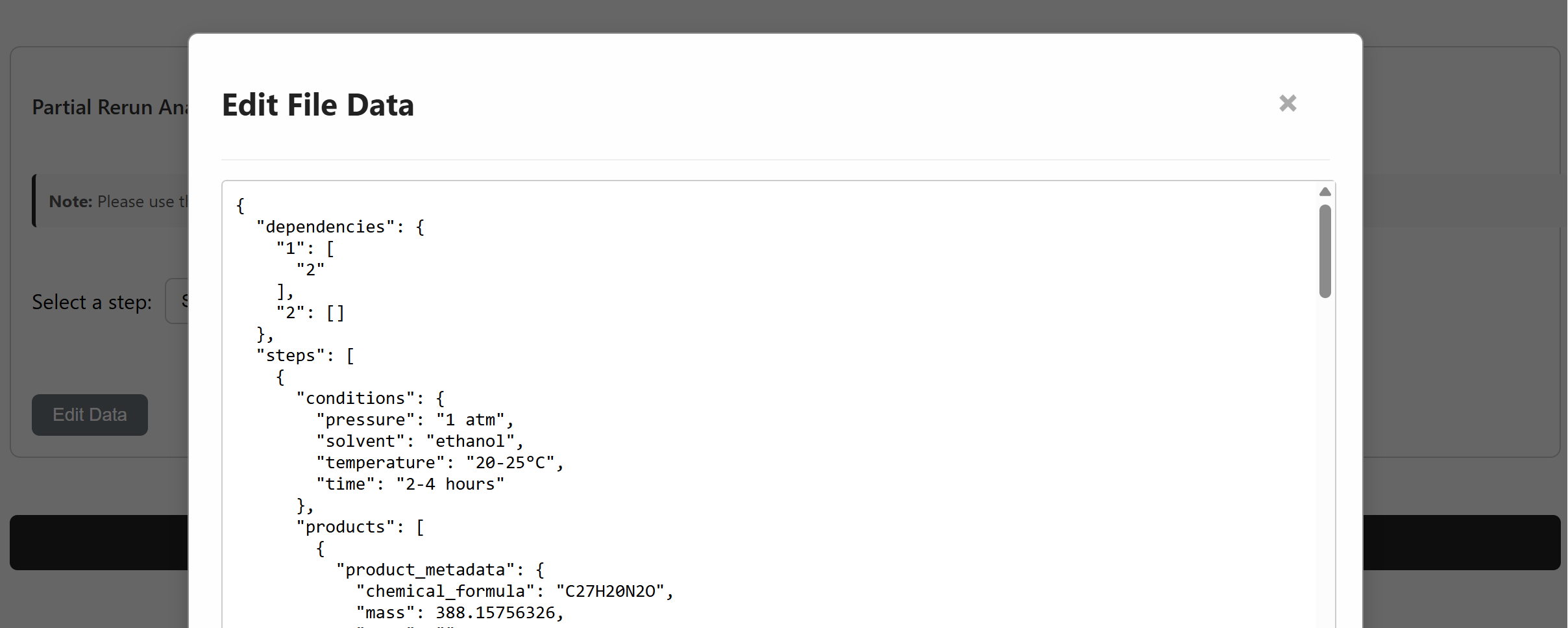
Direct Modification: The interface enables manual condition editing, reagent substitution, and protecting group addition to refine and optimize reaction pathways. Ypu can click on “Edit Data” to directly edit the JSON file and the changes will be automatically reflected in the molecule pathway.
File Management
Pathway File Management
To upload and visualize existing pathways, first click on the “View Pathway” tab in the interface. From there, you can select your JSON file containing the pathway data. The system will automatically load and validate the pathway information. Once loaded, you can easily view the full molecular pathway.
JSON Data Export
To export and manage pathway data, click the “JSON Result” button to view the raw data in JSON format. You can then save this data for further analysis or storage by using the download JSON button provided in the interface.
Multiple Pathway Support

The interface supports handling complex syntheses through pathway switching functionality. You can easily navigate between different proposed routes, compare their efficiency metrics, and evaluate the synthetic complexity of each pathway. This allows you to systematically assess multiple synthetic strategies and select the most promising approach for your target molecule.
Troubleshooting and Best Practices
Troubleshooting
Common Issues and Solutions
API Connection - Solution: Verify server running and API key correct
Verification: Check network and URL configuration
SMILES Errors - Solution: Validate syntax and chemical validity
Verification: Check chemical structure
Visualization Issues - Solution: Refresh page and check browser console
Verification: Cache Clearing
File Upload Problems - Solution: Verify format
Verification: Valid JSON structure
Best Practices
Optimization Guidelines
When working with DeepRetro, it’s important to follow best practices for optimal results. For input validation, start with simple molecules to get familiar with the system. Always verify your SMILES syntax and check chemical validity before proceeding.
For model configuration, ensure you’re using appropriate settings for your specific use case. Enable any relevant validation checks, and optimize the configuration based on the types of molecules you’re working with.
For managing your results:
Review and validate all generated pathways thoroughly
Export any important results for future reference
For advanced usage: User Guide, API Reference, Development Guide